Genomics and the NHS
Personalised and precision medicine
There is currently a shift within medicine towards more personalised treatments, tailoring care specifically to an individual patient. This has been particularly seen in the treatment of various cancers, but is rapidly expanding into other areas. Genomics can play a key role in personalised and precision medicine and has a wide range of potential applications within healthcare including in;
• Risk screening or cascade testing within families for identified genetic conditions (e.g. familial hypercholesterolaemia).
• Personalised treatment - targeted cancer therapies, tailoring dosage or drug choice through pharmacogenomics, avoiding adverse drug reactions.
• Infectious diseases – pathogen identification and outbreak tracing (e.g. tracking variations of COVID-19 strains using RNA sequencing, antimicrobial resistance prediction using whole genome sequencing in Mycobacterium tuberculosis (TB) infection).
• Research – drug design and big data science.
NHS strategy
The NHS has a strategic focus on embedding genomics in practice. In England, this is outlined in the 2022 document Accelerating genomic medicine in the NHS.
• Embedding genomics in NHS from primary/community care through to specialist and tertiary care.
• Delivering equitable genomic testing in cancer, rare, inherited and common diseases, enabling precision medicine and reducing adverse drug reactions.
• Enabling genomics to be at the forefront of the data and digital revolution, ensuring genomic data can be interpreted and informed by other diagnostic and clinical data.
• Evolving the service through cutting-edge science, research and innovation to ensure that patients can benefit from rapid implementation of advances.
• Delivering equitable genomic testing in cancer, rare, inherited and common diseases, enabling precision medicine and reducing adverse drug reactions.
• Enabling genomics to be at the forefront of the data and digital revolution, ensuring genomic data can be interpreted and informed by other diagnostic and clinical data.
• Evolving the service through cutting-edge science, research and innovation to ensure that patients can benefit from rapid implementation of advances.
Pharmacy professionals across all sectors have the potential to significantly contribute to the implementation of personalised medicine and genomics. This could be through helping to implement genomics within the NHS, using evidence-based genomic testing to optimise the prescribing of medicines or reduce toxicity such as in the case studies that follow, or supporting patients in their understanding of the options available to them and aid in shared decision-making. This potential has been recognised by pharmacy leaders as a key area for development and is already incorporated into foundation and advanced frameworks. In April 2023 the Royal Pharmaceutical Society in collaboration with other pharmacy organisations published a position statement on pharmacy and genomics. This describes current and future roles for pharmacy teams in genomic medicine.
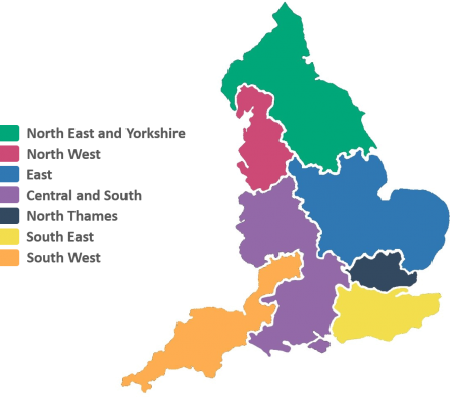 |
| The 7 GMSAs in England Image courtesy of NHS England |
In England, GMSAs bring together research and clinical staff and the regional Genomic Laboratory Hubs as part of the NHS Genomic Medicine Service. Each GMSA has two dedicated pharmacist posts; a Chief Pharmacist for strategic input, and a clinical specialist Consultant Pharmacist for Genomics. If you would like more information on your regional NHS GMS Alliance and their contact details you can e-mail england.genomics@nhs.net
Genomics services in Wales are co-ordinated by Genomics Partnership Wales and the All Wales Medical Genomics Service. The Scottish Strategic Network for Genomic Medicine Laboratory co-ordinates services in Scotland. More information for Northern Ireland Regional Genetics Laboratories – Molecular Genetics can be found on their website.






