Taking and sequencing a genomic sample
Taking a sample
The first step in genomic testing for any individual is acquiring a biological sample, typically blood, saliva or tissue biopsy (the latter particularly for cancer). The sample has the genomic material extracted for testing either for single nucleotide polymorphisms (SNPs) or longer stretches of DNA (for exome or genome sequencing). RNA sequencing is also possible, along with other measurements of cell function (protein synthesis, metabolic activity), although these are less common clinically.
Sequencing a sample
An understanding of the different types of sequencing, when or why they might be used and what they can and can’t be used to identify is helpful when presented with genomic data.
1. Targeted gene panel/SNP analysis
• Only looks at specific genes or SNPs that are known to be associated with a particular disease, phenotype or symptoms (e.g. monogenic diabetes).
• Looking for something you already suspect. Won’t pick up novel causative genes.
• Can’t reanalyse data if new genes are found. A new panel has to be created and retested.
• Exome represents ~2% of human genome, but captures all protein coding regions.
• Majority of disease causing variants are thought to be found in the exome.
• Misses copy number variants or variants outside the exome.
• Can be reanalysed if new genes are identified without need for further sequencing.
• Can be used for ‘virtual targeted panels’ looking for specific gene variants.
3. Whole genome sequencing (WGS)
• Large amount of data requiring costly analysis / storage.
• Can be used for diagnosis, prediction, prevention or treatment of disease.
• WGS can also be used to sequence the genome of tumours and infectious agents to allow effective diagnosis and treatment.
• An example of WGS in practice is the Newborn Genome Programme.
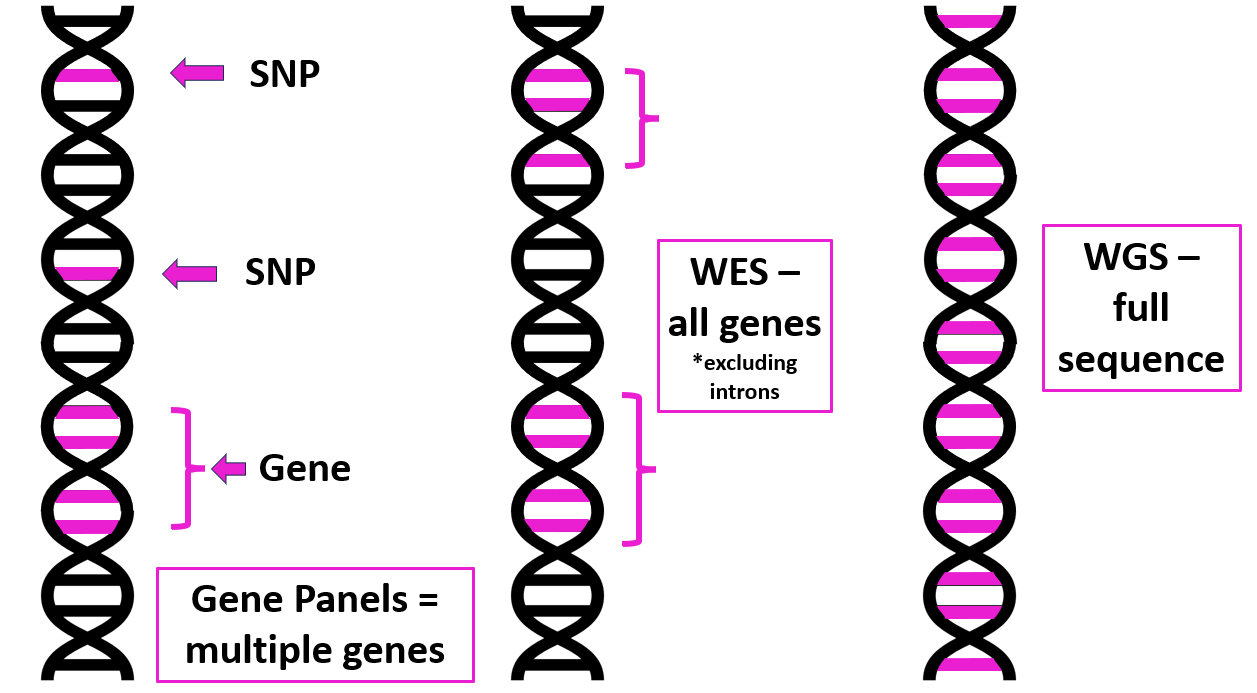 |
| Targeted gene panels, SNP analysis, WES and WGS compared. Image courtesy of Dr Hayley Wickens, Consultant Pharmacist Genomic Medicine |
You can read further information on sequencing on the Genomics Education Programme website.
Examples of genomic sequencing in practice
1. Familial hypercholesterolaemia
Familial hypercholesterolaemia (FH) is a common autosomal dominant genetic disease that increases the likelihood of coronary artery disease, heart attacks and sudden cardiac death at a young age. Early detection and genetic diagnosis can enable early intervention with effective treatments (e.g. statins) and decrease the risk of cardiovascular disease (CVD), leading to better outcomes and reduced morbidity for patients. It can also allow identification and treatment of affected family members.A small targeted panel test is used to identify known gene variants. Patients usually have a functional mutation of one of 3 genes;
• Low-density lipoprotein receptor gene (LDLR or apoB/E receptor) - ~85-90%
• Proprotein convertase subtilisin kexin 9 gene (PCSK9) – 2-4%
• Apolipoprotein B gene (principally APOB3500) – 1-12%
All impair LDL receptor-mediated catabolism resulting in higher LDL-C levels.
Heterozygous FH (person has one copy of a pathogenic variant out of their two copies of the gene) is a common disorder, with an incidence of ~ 1 in 250 individuals in the UK. In England, it is estimated that >150,000 people are affected by FH. Around 7% of those with FH in England have been identified and the aim has been to improve this to 25% by 2024/5. Genetic testing for FH is part of the NHS Long Term Plan (2019).
FH is also currently part of a primary care initiative incentivised by PCN DES IIF 22/23 (CVD-04). This rewards the identification of patients aged 29 and under with a total cholesterol greater than 7.5 mmol/L OR aged 30 and over with a total cholesterol greater than 9.0 mmol/L who have been:
• diagnosed with secondary hyperlipidaemia; or
• clinically assessed for familial hypercholesterolaemia; or
• referred for assessment for familial hypercholesterolaemia; or
• genetically diagnosed with familial hypercholesterolaemia
• clinically assessed for familial hypercholesterolaemia; or
• referred for assessment for familial hypercholesterolaemia; or
• genetically diagnosed with familial hypercholesterolaemia
Pharmacists and pharmacy technicians working in GP practice are ideally placed to facilitate patient identification and prioritisation, counsel and support patients and their families and prescribe and manage therapy.
2. Gene therapy
Another area using genomics and personalised medicine is gene therapy.
The idea behind gene therapy is often to treat the underlying genetic cause of a disease rather than the symptoms. There is potential for gene therapy in areas such as inherited single gene disorders.
One real world example is onasemnogene abeparvovec (Zolgensma) for treating spinal muscular atrophy in babies.
Another application is in cancer therapy. Here a patient’s own white blood cells can be reprogrammed to target their cancer. One example of this, CAR-T (chimeric antigen receptor T-cell) therapy, is already available on the NHS for some cancers.






